Monday, 02 December, 2019
Pattern formation on the cheap
Biophysics

The modular replacements for MinE create this diverse set of patterns when co-reconstituted with MinD on membranes. Credit: Glock et al. (CC BY 4.0)
Many cellular processes involve patterned distributions of proteins. Scientists have identified the minimal set of elements required for the autonomous formation of one such pattern, thus enabling the basic phenomenology to be explored.
Many essential processes in living organisms require the self-organization of specific proteins into precise patterns within cells. For example, autonomous pattern formation is necessary for cell division, active locomotion and intra- and intercellular communication. The Min system in the bacterium Escherichia coli, which is responsible for defining the plane of cell division, is an iconic example of pattern formation. A group led by physicist Erwin Frey (Professor of Statistical and Biological Physics at LMU) and Petra Schwille (Max Planck Institute for Biochemistry) has now identified the minimal set of structural motifs required for the operation of this system. This in turn has allowed them to develop a simplified model with which to investigate the phenomenology of biological pattern formation as a whole. Their findings appear in the online journal eLife.
Cell division in the rod-shaped bacterium E. coli is controlled by two proteins, named MinD and MinE. MinD undergoes modification in the cytoplasm, which causes it to bind to the cell membrane and recruit MinE, thus forming MinDE complexes. After some delay, MinE in turn detaches MinD from the membrane by removing the modification, thus releasing both proteins back into the cytoplasm. This cycle gives rise to intracellular distributions that oscillate back-and-forth in the cell cytoplasm. Ultimately a membrane-bound pattern emerges, which ensures that the plane of cell division is localized to the middle of the cell and not close to either of the poles. Because of its simplicity, this system has provided a valuable model for studying the basic mechanisms that underlie biological formation in general. In addition, the system can be reconstituted from the purified proteins, which allows one to identify the essential functional units and explore the effects of localized mutations on the pattern-forming process.
In the new study, the authors have further simplified this system and identified the minimal set of components required for correct pattern formation. They first created a bare-bones version of MinE by systematically deleting parts of the protein and testing the truncated forms for function. These experiments showed that the short amino-acid sequence which enables MinE to interact with MinD on the membrane and trigger loss of the modification is necessary but not sufficient to implement the process. By successively adding other segments of MinE to this sequence, they obtained several mutants that exhibited the ability to mediate various steps of the process. In addition to its MinD binding sequence, they found that the sequence required for binding of MinE to the cell membrane is also essential for pattern formation. Strikingly, however, analogous membrane-binding sequences from structurally related proteins can be substituted for this sequence.
“Based on these results, we developed a mathematical model that explains why these elements are required for MinE’s function, and how they contribute to pattern formation. In addition, the model predicts how the pattern adapts to the shape of the cell,” says Fridtjof Brauns, who is in Frey’s team and, together with Jacob Halatek (a member of Frey’s group) and Philipp Glock, a PhD student at the MPI for Biochemistry, joint first author of the new paper.“With this model, one can now ask, irrespective of the specific protein system considered, what functional features must be present in order to make self-organization and pattern formation possible,” says Frey. The new study thus constitutes a significant advance in the quest for a comprehensive understanding of protein-based pattern formation in biological systems. 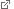 eLife 2019
eLife 2019
Source:  LMU press release
LMU press release

