Thursday, 18 April, 2019
Multiple modes for selectivity of transmembrane transport
Molecular Biophysics

Closed conformation of a bacterial substrate-binding protein following the binding of a substrate. Source: T. Cordes
LMU researchers utilized a biophysical approach to understand how bacterial import proteins bind and selectively convey their cargoes across membranes. The results reveal an unexpectedly wide variety of transfer mechanisms.
Many essential cellular functions rely on the selective uptake or export of specific cargoes across membranes. These vital tasks are performed by specialized, membrane-bound, transport proteins. Transporters of this type are required for the import of nutrients from the surrounding medium, the disposal of toxic products or the defense of pathogens. As in any import-export business, the most important aspect of these transport processes is the selection of the appropriate substrates. LMU biophysicist Professor Thorben Cordes and his team have examined how representative members of an important class of transport proteins called bacterial ABC importers select their preferred cargoes. They identified a wide range of binding mechanisms, which are largely dependent on the fact that one domain of the transporters exhibits a surprising degree of structural flexibility. The new findings appear in the online journal eLife.
Transport by bacterial ABC import systems begins with the recognition of a potential cargo molecule by a specific substrate binding protein (SBP). Successful binding alters the three-dimensional structure of the binding protein from an open to a closed conformation. It was generally assumed that the conformational change induced by substrate binding dictates the selectivity of substrate transport by enabling the loaded SBP to interact with the transporter subunit, which is responsible for conveying the substrate across the membrane. “However, there were various indications that some molecules bind the SBPs with high affinity but are not subsequently transported across the membrane,” says Cordes. “To explore the relationship between binding and transport, we used a spectroscopic method that characterizes both structural changes and dyamics caused by the binding of different substrates, and asked whether these contribute to the selectivity of substrate uptake by bacterial transport systems.“
The methodology applied by Cordes and his colleagues Bert Poolman (Groningen, NL) and Christopher McDevitt (Melbourne, AUS) differs from the classical structural biology approaches, which aim at the determination of the three-dimensional structure of the entire proteins, with and without their binding partners. Instead, they used a technique known as single-molecule fluorescence energy transfer (smFRET). This effectively allows one to measure the distance between two fluorescent dyes that have been chemically attached to specific positions in a protein. The method therefore makes it possible to monitor shifts in the conformational states of SBPs, and has the great advantage that the interactions of many different substrates with the same SBP can be rapidly characterized. With the aid of this approach, it was shown that the SBPs found in ABC systems are surprisingly flexible. “There is no such thing as a single, clearly definable, closed conformation that activates transport,” says Cordes. Instead, binding proteins are capable of adopting a range of activating and non-activating conformations.”
In some cases, binding of transported substrates and non-transported substrates results in very similar conformations. In such cases, factors other than the conformation of the binding protein must determine the selectivity of the transport step. “Our results reveal an unexpected diversity of transport selection mechanisms in these proteins,” says Cordes. “We hope that this insight will contribute to a fundamental understanding of the mechanisms utilized by ABC transporters. Such an understanding could also be of medical interest, as the binding proteins we have looked at are specific to bacterial cells. On the basis of our findings, it might therefore be possible to develop novel antibiotics.”
eLife 2019
For further information, see:
Molecular machines at work
Source: 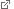 LMU press release
LMU press release LMU Pressemitteilung auf Deutsch
LMU Pressemitteilung auf Deutsch

